Recent technology advancements have catalyzed the rapid growth of large volumes of biomedical data with complex structures — particularly medical imaging data. The sheer volume and relatively unstructured nature of imaging data, coupled with its ability to carry significant amounts of information on disease status and treatment outcomes, exemplify the double-edged sword of big data for the medical world. Although more data is being collected to develop sophisticated statistical models and to train and test machine learning algorithms, the amount of useful information extracted is not necessarily proportional to the size of the data.
Large-scale data can be noisy, heterogeneous, highly correlated and non-informative for the targeted scientific problems. This has created a high demand for developing innovative statistical methods and computational scalable algorithms that can extract critical information. Chancellor’s Professor of Statistics Annie Qu and her team, working to address this demand, are developing methods and tools to enhance the detection of early stage invasive cancers using medical imaging data.
Advanced Imaging Requires Advanced Analytics
Common breast cancer imaging techniques, such as mammography, are not able to differentiate high-risk breast cancer from low-risk breast cancer cases, which leads to inaccurate diagnosis and prognosis, and prevents potential invasive cancer patients from receiving early cancer intervention and treatment. Motivated by multimodal multiphoton optical breast cancer imaging data produced by the Boppart Lab at the University of Illinois at Urbana-Champaign and the Carle Foundation Hospital in Illinois, Professor Qu is exploring how to leverage optical imaging data produced by the newly developed multiphoton microscopes.
These microscopes generate multimodal images through multiphoton auto-fluorescence and multiharmonic generation. The new technique is able to identify cancer cell clusters in a specimen that are not easily identified by histology imaging, which has low contrast to visualize cell bodies or vesicles under microenvironments.
In contrast to histology or mammographic images, multiphoton images enable more comprehensive cancer diagnosis or prognosis obtained from the unperturbed tumor microenvironment without external labeling or stains. For example, Figure 1 illustrates four modalities for normal human breast tissue and for tumor breast specimens, showing the clear concentration of tumor-associated microvesicles (TMVs) as observed in certain modality images from tumor samples but not obvious from normal samples.
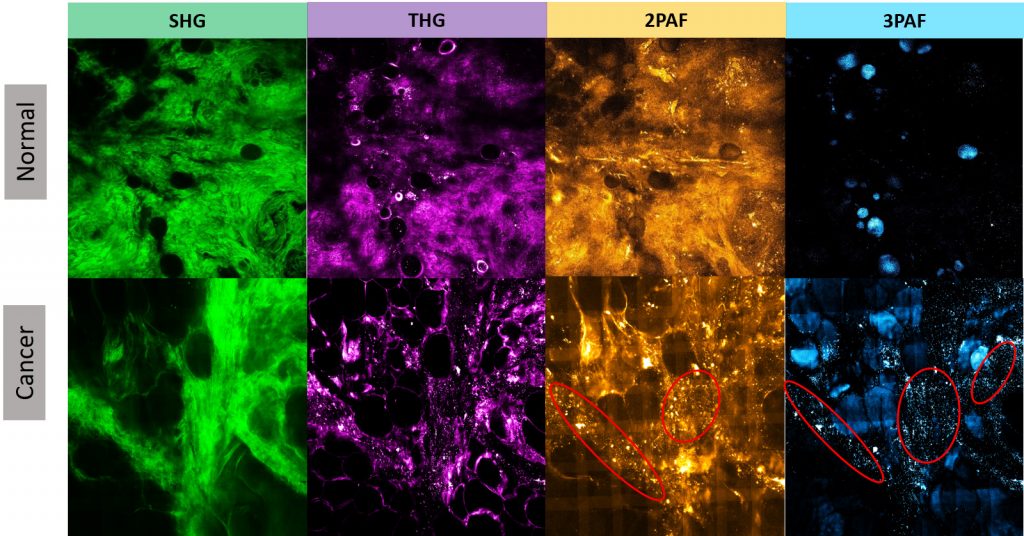
The microvesicles play an important role in understanding the development of carcinogenesis prior to metastatic infiltration. The TMVs are established as a specific biomarker that can precede the appearance of invasive tumors, as large numbers of microvesicles are highly correlated with invasive tumor cell growth. In addition, microvesicles carry important molecular and genetic information from tumors and affect several stages in tumor progression.
The advanced multimodality multiphoton imaging provides a visualization of the spatial dynamics of TMVs. However, critical information from such spatial dynamics is not fully utilized in current imaging data analyses. Developing cutting-edge statistical methods and machine learning tools is urgently needed to fully exploit the wealth of medical imaging data.
Innovations in Diagnostic Methods
One unique challenge for optical imaging is that the microvesicle signal strength is relatively weak compared to the modality-specific background information. Indeed, a typical deep learning method is not effective in detecting TMVs in optical imaging data. This is not surprising, given that the breast cancer optical images from individual patients present highly heterogenous spatial patterns associated with microvesicles. Although the number of patients is large, the number of imaging modalities from the same patient is still limited.
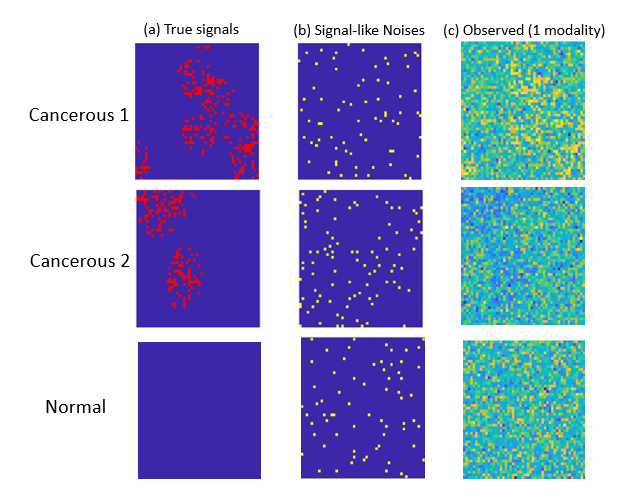
Professor Qu and her students have developed new diagnostic methods that integrate correlated multimodality and spatial information of signals simultaneously. Their proposed individualized multilayer tensor learning (IMTL) method uses additional individualized layers to incorporate heterogeneity into a higher-order tensor decomposition (a sum of rank-1 outer products of multiple vectors). They also extended their method to integrate individual-specific information over multimodalities and to incorporate modality-specific features through different layers.
From a feature-extraction perspective, the decomposed basis tensor layers in the proposed approach can be viewed as some full-size feature extraction filters analogous to those in the convolution neural network (CNN). However, in contrast to the CNN, which applies homogeneous filters over all subjects, the individualized layers serve as subject-specific filters to capture individual-wise characteristics and heterogeneity information, which can effectively capture heterogeneous outcomes such as the TMVs in breast cancer imaging data.
Professor Qu and her team have also introduced the concept of “correlation tensor” to represent spatial correlations from multimodal imaging. One advantage of the correlation tensor is that it can represent the correlation structures jointly for multimodal data and capture spatial information more effectively. In addition, the team has proposed a semi-symmetric tensor decomposition method to effectively extract the true signal from the noisy modality-specific background of optical imaging. This leads to highly efficient estimation and accurate classification when the number of parameters exceeds the sample size. The proposed method is especially effective when the signals of interest are weak compared to noisy background information.
By integrating cutting-edge optical imaging techniques, statistical modeling, machine learning, and computational tools, Professor Qu and her team are working to improve the detection for invasive cancers in microenvironments, reducing cancer mortality rates and minimizing medical costs in the future.